class: center, middle # An Overview of R - 2 ## Introductory Computer Programming ### Deepayan Sarkar <h1 onclick="document.documentElement.requestFullscreen();" style="cursor: pointer;"> <svg xmlns="http://www.w3.org/2000/svg" width="16" height="16" fill="currentColor" class="bi bi-arrows-fullscreen" viewBox="0 0 16 16"> <path fill-rule="evenodd" d="M5.828 10.172a.5.5 0 0 0-.707 0l-4.096 4.096V11.5a.5.5 0 0 0-1 0v3.975a.5.5 0 0 0 .5.5H4.5a.5.5 0 0 0 0-1H1.732l4.096-4.096a.5.5 0 0 0 0-.707zm4.344 0a.5.5 0 0 1 .707 0l4.096 4.096V11.5a.5.5 0 1 1 1 0v3.975a.5.5 0 0 1-.5.5H11.5a.5.5 0 0 1 0-1h2.768l-4.096-4.096a.5.5 0 0 1 0-.707zm0-4.344a.5.5 0 0 0 .707 0l4.096-4.096V4.5a.5.5 0 1 0 1 0V.525a.5.5 0 0 0-.5-.5H11.5a.5.5 0 0 0 0 1h2.768l-4.096 4.096a.5.5 0 0 0 0 .707zm-4.344 0a.5.5 0 0 1-.707 0L1.025 1.732V4.5a.5.5 0 0 1-1 0V.525a.5.5 0 0 1 .5-.5H4.5a.5.5 0 0 1 0 1H1.732l4.096 4.096a.5.5 0 0 1 0 .707z"/> </svg> </h1> --- # Before we start, an experiment! <!-- https://developer.mozilla.org/en-US/docs/Web/CSS/cursor --> <p><img src="the_dress.png" style="width:25%;" alt="the dress" /><br /> <!--  --> -- Color combination: Is it __white & gold__ or __blue & black__ ? Let's count! ```r NN <- 18 + 8 # total XX <- 18 # white and gold ``` --- # Question: What proportion of population sees white & gold? - Statistics uses data to make inferences - Model: - Let $p$ be the probability of seeing white & gold - Assume that individuals are independent -- - Data: - Suppose $X$ out of $N$ sampled individuals see white & gold; e.g., $N = 26$, $X = 18$. - According to model, $X \sim Bin(N, p)$ -- - "Obvious" estimate of $p = X / N = 18 / 26 = 0.6923$ - But how is this estimate derived? --- # Generally useful method: maximum likelihood - Likelihood function: probability of observed data as function of $p$ $$ L(p) = P(X = 18) = {26 \choose 18} p^{18} (1-p)^{(26-18)}, p \in (0, 1) $$ - Intuition: $p$ that gives higher $L(p)$ is more "likely" to be correct - Maximum likelihood estimate $\hat{p} = \arg \max L(p)$ -- - By differentiating $$ \log L(p) = c + 18 \log p + 8 \log (1-p) $$ we get $$ \frac{d}{dp} \log L(p) = \frac{18}{p} - \frac{8}{1-p} = 0 \implies 18 (1-p) - 8 p = 0 \implies p = \frac{18}{26} $$ --- # How could we do this numerically? - Pretend for the moment that we did not know how to do this. - How could we arrive at the same solution numerically? - Basic idea: Compute $L(p)$ for various values of $p$ and find minimum. -- - To do this in R, remember that R works like a calculator: - The user types in an expression, R calculates the answer - The expression can involve numbers, variables, and functions - For example: ```r N = NN x = XX p = 0.5 choose(N, x) * p^x * (1-p)^(N-x) ``` ``` [1] 0.02327971 ``` --- # "Vectorized" computations - One important distinguishing feature of R is that it operates on "vectors" ```r pvec = seq(0, 1, by = 0.01) pvec ``` ``` [1] 0.00 0.01 0.02 0.03 0.04 0.05 0.06 0.07 0.08 0.09 0.10 0.11 0.12 0.13 0.14 0.15 0.16 0.17 0.18 [20] 0.19 0.20 0.21 0.22 0.23 0.24 0.25 0.26 0.27 0.28 0.29 0.30 0.31 0.32 0.33 0.34 0.35 0.36 0.37 [39] 0.38 0.39 0.40 0.41 0.42 0.43 0.44 0.45 0.46 0.47 0.48 0.49 0.50 0.51 0.52 0.53 0.54 0.55 0.56 [58] 0.57 0.58 0.59 0.60 0.61 0.62 0.63 0.64 0.65 0.66 0.67 0.68 0.69 0.70 0.71 0.72 0.73 0.74 0.75 [77] 0.76 0.77 0.78 0.79 0.80 0.81 0.82 0.83 0.84 0.85 0.86 0.87 0.88 0.89 0.90 0.91 0.92 0.93 0.94 [96] 0.95 0.96 0.97 0.98 0.99 1.00 ``` ```r Lvec = choose(N, x) * pvec^x * (1-pvec)^(N-x) Lvec ``` ``` [1] 0.000000e+00 1.441581e-30 3.484224e-25 4.743664e-22 7.744746e-20 3.953724e-18 9.671700e-17 [8] 1.423593e-15 1.444375e-14 1.102691e-13 6.725082e-13 3.419368e-12 1.495827e-11 5.766228e-11 [15] 1.995491e-10 6.291462e-10 1.828738e-09 4.948377e-09 1.256532e-08 3.014411e-08 6.870958e-08 [22] 1.495277e-07 3.119770e-07 6.263028e-07 1.213580e-06 2.275976e-06 4.141251e-06 7.326358e-06 [29] 1.262589e-05 2.123187e-05 3.489188e-05 5.611264e-05 8.841550e-05 1.366491e-04 2.073621e-04 [36] 3.092354e-04 4.535679e-04 6.548064e-04 9.310987e-04 1.304845e-03 1.803214e-03 2.458572e-03 [43] 3.308786e-03 4.397325e-03 5.773111e-03 7.490042e-03 9.606132e-03 1.218220e-02 1.528011e-02 [50] 1.896044e-02 2.327971e-02 2.828716e-02 3.402104e-02 4.050475e-02 4.774279e-02 5.571678e-02 [57] 6.438184e-02 7.366353e-02 8.345564e-02 9.361912e-02 1.039824e-01 1.143435e-01 1.244734e-01 [64] 1.341224e-01 1.430267e-01 1.509184e-01 1.575359e-01 1.626355e-01 1.660037e-01 1.674691e-01 [71] 1.669138e-01 1.642827e-01 1.595904e-01 1.529252e-01 1.444492e-01 1.343941e-01 1.230530e-01 [78] 1.107677e-01 9.791332e-02 8.487885e-02 7.204722e-02 5.977459e-02 4.837120e-02 3.808505e-02 [85] 2.909005e-02 2.147970e-02 1.526701e-02 1.039084e-02 6.728148e-03 4.110731e-03 2.344891e-03 [92] 1.231523e-03 5.843301e-04 2.439130e-04 8.615269e-05 2.424055e-05 4.910431e-06 5.924050e-07 [99] 2.780141e-08 1.303740e-10 0.000000e+00 ``` --- # Plotting is very easy ```r plot(x = pvec, y = Lvec, type = "l") ``` 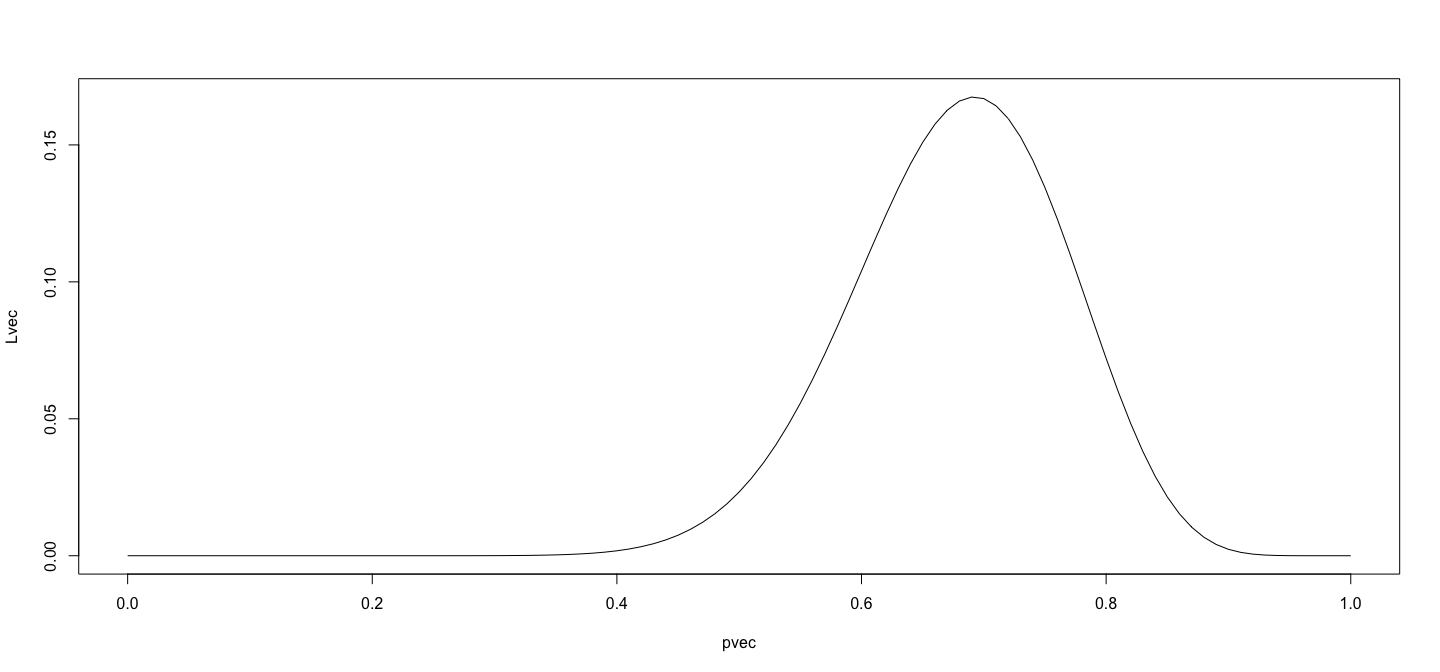 --- # Functions - Functions can be used to encapsulate repetitive computations - Like mathematical functions, they take arguments as input and "returns" an output ```r L = function(p) choose(N, x) * p^x * (1-p)^(N-x) L(0.5) ``` ``` [1] 0.02327971 ``` ```r L(x/N) ``` ``` [1] 0.1675234 ``` --- # Functions can be plotted directly ```r plot(L, from = 0, to = 1) ``` 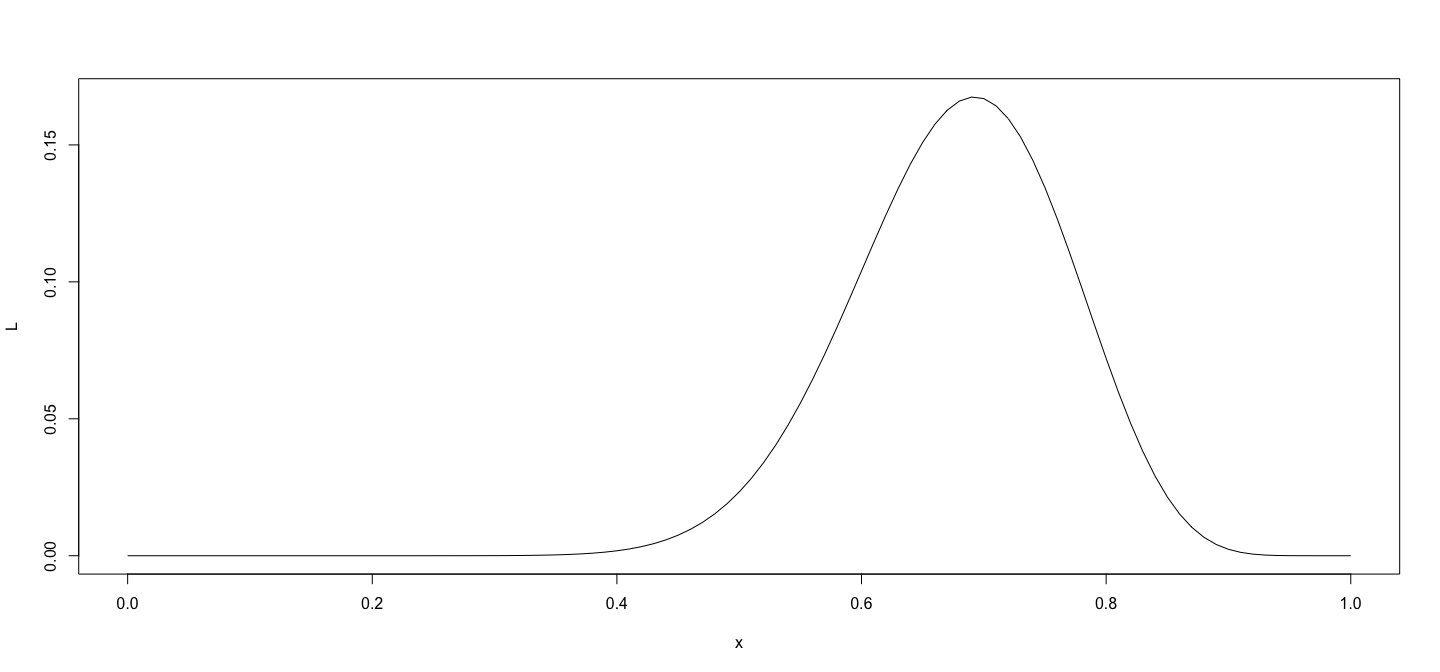 --- # ...and they can be numerically "optimized" ```r optimize(L, interval = c(0, 1), maximum = TRUE) ``` ``` $maximum [1] 0.6923109 $objective [1] 0.1675234 ``` - Compare with ```r XX / NN ``` ``` [1] 0.6923077 ``` --- # Another example: Linear regression ```r data(Davis, package = "carData") # Davis height and weight data str(Davis) # Summarize structure of data ``` ``` 'data.frame': 200 obs. of 5 variables: $ sex : Factor w/ 2 levels "F","M": 2 1 1 2 1 2 2 2 2 2 ... $ weight: int 77 58 53 68 59 76 76 69 71 65 ... $ height: int 182 161 161 177 157 170 167 186 178 171 ... $ repwt : int 77 51 54 70 59 76 77 73 71 64 ... $ repht : int 180 159 158 175 155 165 165 180 175 170 ... ``` ```r fm <- lm(weight ~ height, data = Davis) # Regression of weight on height fm ``` ``` Call: lm(formula = weight ~ height, data = Davis) Coefficients: (Intercept) height 25.2662 0.2384 ``` --- # You should always plot the data first! ```r library(lattice) # A different way to plot data xyplot(weight ~ height, data = Davis, grid = TRUE, type = c("p", "r")) ``` 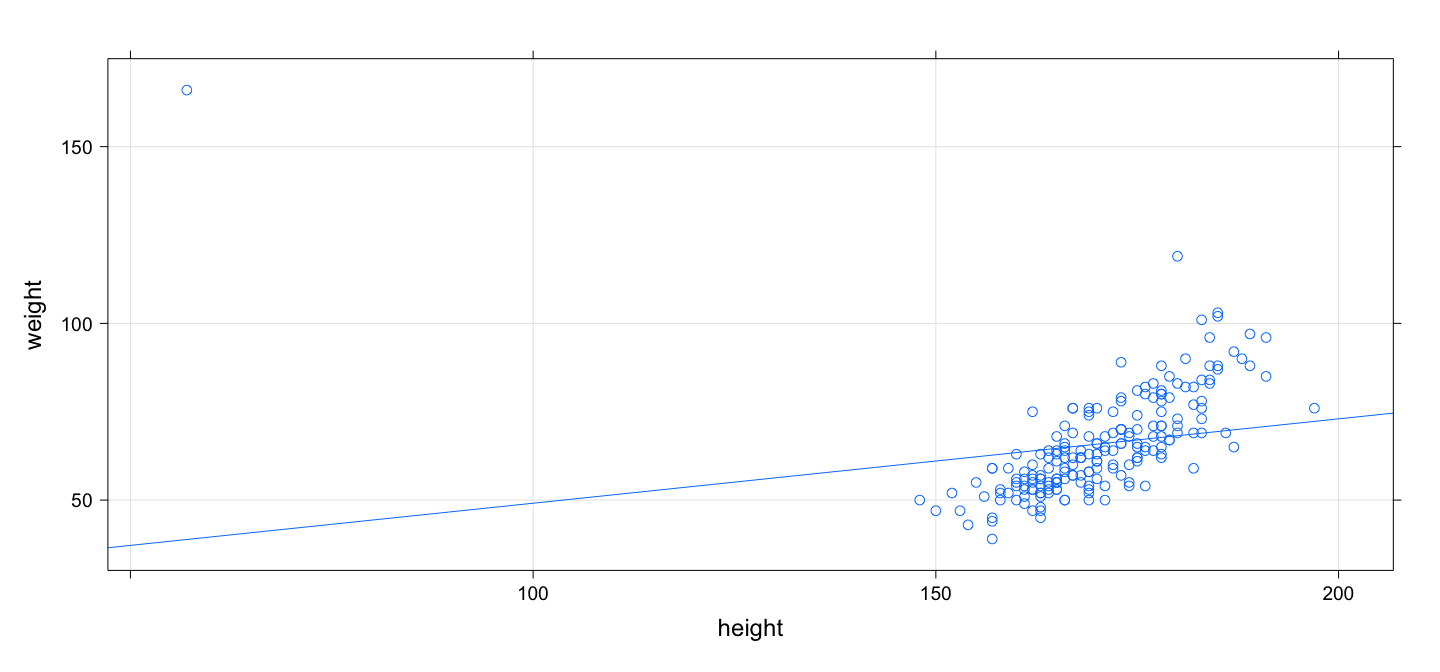 --- # The regression model is fit by minimizing least squares ```r coef(fm) # estimated regression coefficients ``` ``` (Intercept) height 25.2662278 0.2384059 ``` -- - We can confirm using a general optimizer: ```r SSE = function(beta) { with(Davis, sum((weight - beta[1] - beta[2] * height)^2)) } optim(c(0, 0), fn = SSE) ``` ``` $par [1] 25.3053648 0.2381936 $value [1] 43713.12 $counts function gradient 99 NA $convergence [1] 0 $message NULL ``` --- # Fitting the regression model - `lm()` gives exact solution and more statistically relevant details ```r summary(fm) ``` ``` Call: lm(formula = weight ~ height, data = Davis) Residuals: Min 1Q Median 3Q Max -23.696 -9.506 -2.818 6.372 127.145 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 25.26623 14.95042 1.690 0.09260 . height 0.23841 0.08772 2.718 0.00715 ** --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 14.86 on 198 degrees of freedom Multiple R-squared: 0.03597, Adjusted R-squared: 0.0311 F-statistic: 7.387 on 1 and 198 DF, p-value: 0.007152 ``` --- # Changing the model-fitting criteria - Suppose we wanted to minimize _sum of absolute errors_ instead of sum of squares - No closed form solution any more, but general optimizer will still work: ```r SAE = function(beta) { with(Davis, sum(abs(weight - beta[1] - beta[2] * height))) } opt = optim(c(0, 0), fn = SAE) opt ``` ``` $par [1] -106.000787 1.000005 $value [1] 1504 $counts function gradient 169 NA $convergence [1] 0 $message NULL ``` --- # This is an example of _robust regression_ ```r xyplot(weight ~ height, data = Davis, grid = TRUE, panel = function(x, y, ...) { panel.abline(fm, col = "red") # squared errors panel.abline(opt$par, col = "blue") # absolute errors panel.xyplot(x, y, ...) }) ``` 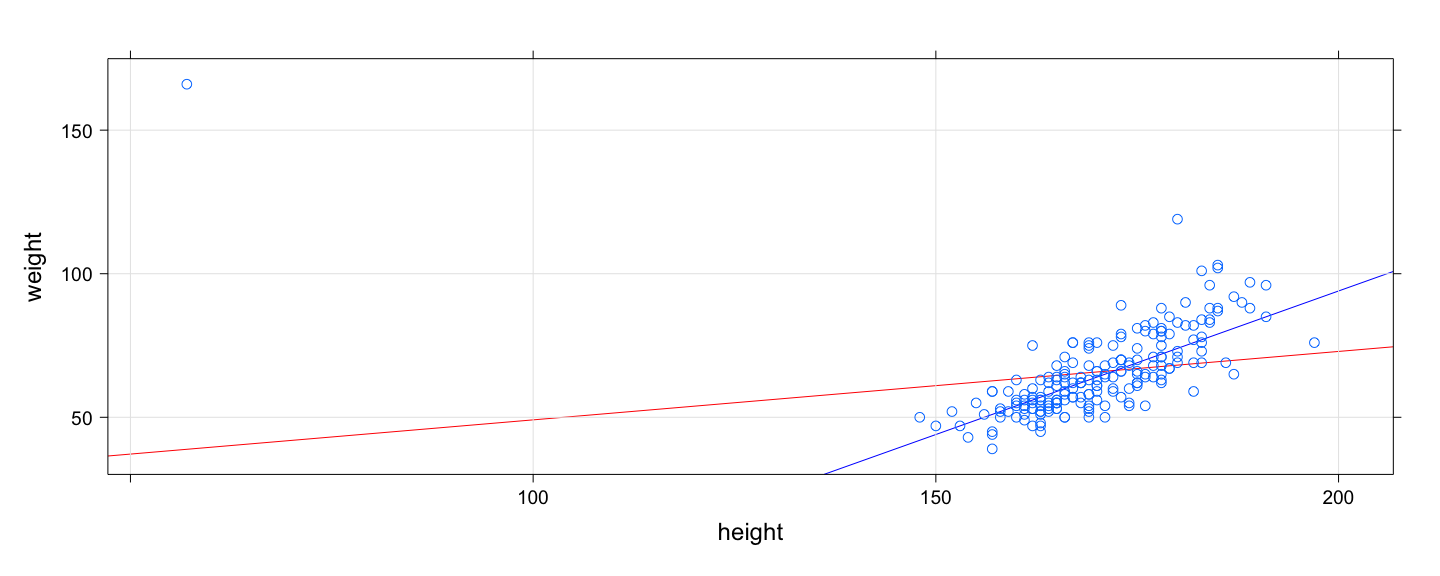 --- # Another way to plot the data and model fits ```r library(ggplot2) qplot(x = height, y = weight, data = Davis) + geom_abline(intercept = coef(fm)[1], slope = coef(fm)[2], col = 2) + geom_abline(intercept = opt$par[1], slope = opt$par[2], col = 3) ```  --- # With an easy way to make the plot interactive ```r library(plotly) ggplotly(qplot(x = height, y = weight, data = Davis) + geom_abline(intercept = coef(fm)[1], slope = coef(fm)[2], col = 2) + geom_abline(intercept = opt$par[1], slope = opt$par[2], col = 3)) ``` <iframe src="davis-plotly.html" height="400px" width="99%"></iframe> --- # Test for slope * Natural follow up question: Is slope non-zero? * Need a test statistic * Null distribution should not depend on $\sigma^2$ * How can we find one? -- * Suppose we somehow obtain a test statistic * How can we obtain the corresponding $p$-value? --- # Simulation * R can be used to simulate random events * Example: how likely is a common birthday in a group of 20 people? ```r N <- 28 days <- sample(365, N, rep = TRUE) days ``` ``` [1] 112 66 311 109 161 47 183 276 210 255 90 146 207 229 298 75 294 358 340 253 21 137 219 256 [25] 83 278 179 172 ``` ```r length(unique(days)) ``` ``` [1] 28 ``` --- layout: true # Law of Large Numbers * With enough replications, sample proportion should converge to probability (consistency) --- ```r haveCommon <- function() { days = sample(365, N, rep = TRUE) length(unique(days)) < N } haveCommon() ``` ``` [1] FALSE ``` ```r haveCommon() ``` ``` [1] TRUE ``` ```r haveCommon() ``` ``` [1] FALSE ``` ```r haveCommon() ``` ``` [1] FALSE ``` --- * Do this sytematically: ```r replicate(100, haveCommon()) ``` ``` [1] FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE FALSE FALSE TRUE FALSE FALSE TRUE FALSE [16] TRUE TRUE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE TRUE TRUE [31] FALSE FALSE TRUE FALSE FALSE FALSE FALSE TRUE FALSE FALSE TRUE TRUE FALSE TRUE TRUE [46] TRUE TRUE TRUE TRUE TRUE FALSE TRUE TRUE TRUE FALSE TRUE TRUE FALSE TRUE TRUE [61] TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE TRUE TRUE FALSE FALSE [76] TRUE TRUE TRUE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE [91] TRUE TRUE TRUE FALSE TRUE TRUE TRUE TRUE TRUE TRUE ``` --- ```r plot(cumsum(replicate(1000, haveCommon())) / 1:1000, type = "l", ylim = c(0.5, 1)) lines(cumsum(replicate(1000, haveCommon())) / 1:1000, col = "red") lines(cumsum(replicate(1000, haveCommon())) / 1:1000, col = "blue") ```  --- layout: false # R gives access to an extensive toolset * Most standard data analysis methods are already implemented * Can be extended by writing add-on packages * Thousands of add-on packages are available - The last few plots are all created using add-on packages --- # Drawbacks * Learning R needs some effort * Not point-and-click software * Command-line interface --- # Good practices * Use a good interface (R Studio is the most popular one) * Save your code in a script file (makes it easier to reproduce later) * Use "literate programming" approaches (notebooks, R Markdown, etc.) * Demo using R using [R script](https://github.com/deepayan/deepayan.github.io/blob/master/courses/introductory-computer-programming/slides/03-roverview-2.R) and an [R Markdown](https://github.com/deepayan/deepayan.github.io/blob/master/courses/introductory-computer-programming/slides/03-roverview-2.rmd) file